Bacteriophages – viruses that infect bacterial cells – were almost forgotten in the age of antibiotics. Now as bacterial resistance grows, they may return to help us in our hour of need.
There are about 1,400 identified human pathogens in the world. Many are bacterial species, which in turn can be divided into thousands of different strains. Unfortunately, many have genetic mutations that make them resistant to our primary weapon against them: antibiotics. As these resistant strains have emerged, this weapon has weakened over time, and developing new antibiotics has been tricky. But even if we are not able to find new antibiotics to fend off resistant bacteria, we can still win the war. One way to do so is to work with bacteria’s natural enemy: the bacteriophage.
The end of the golden age of antibiotics
Antibiotic resistance has now been recognized as one of the biggest threats to global health. Globally, as many as 700,000 people die every year due to resistant bacterial infections, including thousands in developed countries with otherwise adequate healthcare systems. One estimate predicts that there will be millions of annual deaths from resistant bacteria by 2050, if things continue on the trajectory they’re on today.
Antibiotics are small molecules that have the ability to restrain the growth of bacteria or outright kill them. Initially, the term antibiotics referred to natural compounds produced by microorganisms that killed or inhibited other microorganisms which were competing for nutrients and space in a shared environmental niche. But nowadays, the term also includes semi-synthetic or synthetic compounds used in medicine.
Antibiotics are mostly safe for humans – given the right dose – as they target specific structures that are only found in bacterial cells, but not in the cells of humans or other organisms, such as animals, plants or fungi. But due to this specificity, they can only be used to treat bacterial infections and are useless against viral infections, like a common cold or the flu.
Their specificity also distinguishes them from other chemicals like disinfectants or detergents, which have a much broader mode of action targeting many cells in many organisms. Although these are useful to remove bacteria from surfaces, such as your hands or a kitchen counter, they would not be suited to treat an infection inside an organism, because they would also be toxic towards our cells if we ingested them or injected them into our bodies.
When Alexander Fleming discovered the first antibiotic – penicillin – in 1928, his discovery was followed by a golden age of antibiotics, which peaked between 1940 and the late 1960s. Remarkably, half of the antibiotics we have available today were discovered during this period. Suddenly, many life-threatening diseases became very easy to treat. Surgeries that once risked fatal infections became safe.
But since that time, the misuse and overuse of antibiotics has dramatically accelerated the occurrence of resistant bacteria.
Although resistance against antibiotics can be observed in nature, the mechanisms that lead to resistance development act much more slowly in nature than under the man-made pressure of antibiotics.
Indeed, bacteria develop resistance quickest when antibiotics are misused – whether through over- or under-use. When bacteria are exposed to antibiotics at a dose high enough to kill the whole population, the infection will be cleared very efficiently. But there is a chance that a tiny fraction of the group has a rare mutation that makes it harder for a given antibiotic to kill them. With nearly every bacterium in a given bacterial population wiped out, the surviving fraction can quickly multiply and rebuild the population – now resistant to the drug.
On the other hand, if antibiotic treatment is stopped too early or the dose is not high enough to wipe out the entire bacterial population, the bacteria gain time to adapt. The ones that acquire beneficial mutations that allow them to survive and grow under the antibiotic pressure will outcompete the others and eventually the whole population will become resistant.
The ‘strategies’ that bacteria ‘use’ – i.e. the mutations that get selected for by antibiotic use – are manyfold. First, they can restrict the access of the antibiotic by changing the entryway, e.g. by changing their surface such that the antibiotic can no longer enter them. Second, they can actively pump antibiotics out of the cell. Or they can deactivate the antibiotic by destroying it or changing its chemical characteristics.
Sometimes, bacteria gain mutations that help them actually change the targets of those antibiotics in the cell or bypass their effects by developing new cellular processes. A well-known example is the way that Staphylococcus aureus developed resistance against β-lactam antibiotics, the class of antibiotics that includes penicillin. β-lactam antibiotics interfere with the construction of the cell wall of bacteria, by binding to a protein called penicillin binding protein (PBP) which is crucial in the synthesis of the cell wall. When β-lactam antibiotics bind to this protein, they prevent it from properly structuring the cell wall. Pressure rises inside bacterial cells and they burst. But resistant strains have evolved a new PBP with a different structure, which makes it impossible for the antibiotic to bind its target.
Resistant bacteria can divide and spread among humans and in the environment, and they can even spread those resistance mechanisms to entirely different species of bacteria.
Bacterial cells can transfer genetic material between them through three main mechanisms. When bacterial cells contact each other directly, it can happen that they transfer their genetic material. Free DNA can also be taken up through a process called transformation. The third way is through phages. Phages can transfer DNA from a previously infected host to a new host.
If a first-line antibiotic such as amoxicillin fails to cure your infection, and your immune system can’t handle it either, then hospitals draw on second- and third-line antibiotics. However, this does not come without risks. There is the aforementioned danger that resistance to the back-up antibiotics spreads further, making them less effective in the future. Also, such antibiotics can have worse side effects even if used carefully.
Clearly, we need more treatments, but our pipeline of excellent antibiotics seems to have slowed down after the initial flurry. If we picture the battle against bacteria as a tug of war we pulled the rope hugely in our direction with the invention of penicillin, and kept it that way for a few decades. But since then we have lost ground. We’re still winning, but not as decisively – and the direction of travel is negative.
Antibiotic resistance leads to higher mortality, longer hospital stays and higher healthcare costs. It is becoming more and more apparent that antibiotics alone are not enough to tackle this problem. This desperate need for new approaches has led to the rediscovery of an almost forgotten option: phage therapy.
Enter: Bacteriophages
Bacteriophages – or phages for short – are the natural enemy of bacteria. They are viruses which infect bacteria to reproduce. The word phage originated from the ancient Greek word ‘φαγεῖν’ (phagein), which literally means ‘to eat’ or ‘to devour’.
Phages were originally discovered at the beginning of the 20th century by both Frederick Twort, and Félix d’Hérelle, independently. Twort described ‘glassy and transparent’ spots on plates containing bacterial cultures. Those spots turned out to be dead bacteria. What was crucial about the discovery was that he observed that those spots could be transmitted. He diluted material from the spots in water to a concentration of one in a million and filtered it. With just a single drop onto another fresh plate that contained a bacterial culture, he found the formation of a glassy spot there as well. He noticed that those spots could not grow on their own or on different bacterial cultures, but that they required the same species of bacterial culture to grow. He then suggested that the cause of those glassy spots were ‘ultra-microscopic viruses’.
Over time, with development of new technologies like electron microscopy, scientists could confirm the presence of these ‘bacteriophages’. Like other viruses, they consist of proteins that encapsulate their genetic material. Being viruses, they cannot reproduce on their own, but do so as parasites, inside other organisms. In their case, those other organisms are bacteria. Most phages are quite specific. Some can only reproduce in one particular strain of one species, but all of them are limited to reproduction in bacterial cells. This makes them harmless to humans, animals and plants.
Phages interact with a specific receptor on the cell surface of their target bacterium. Like having a key for a specific lock, they can attach to particular bacteria and inject their genetic material into the cell. Inside the cell, the phage can start its ‘lytic cycle’, where it immediately hijacks the bacterium to produce copies of itself. Or it can enter the lysogenic cycle where it goes dormant, keeping its genetic material inside its host, as a “prophage”, until later on. If its host bacteria is under stress, this takes it out of dormancy and triggers the lytic cycle.
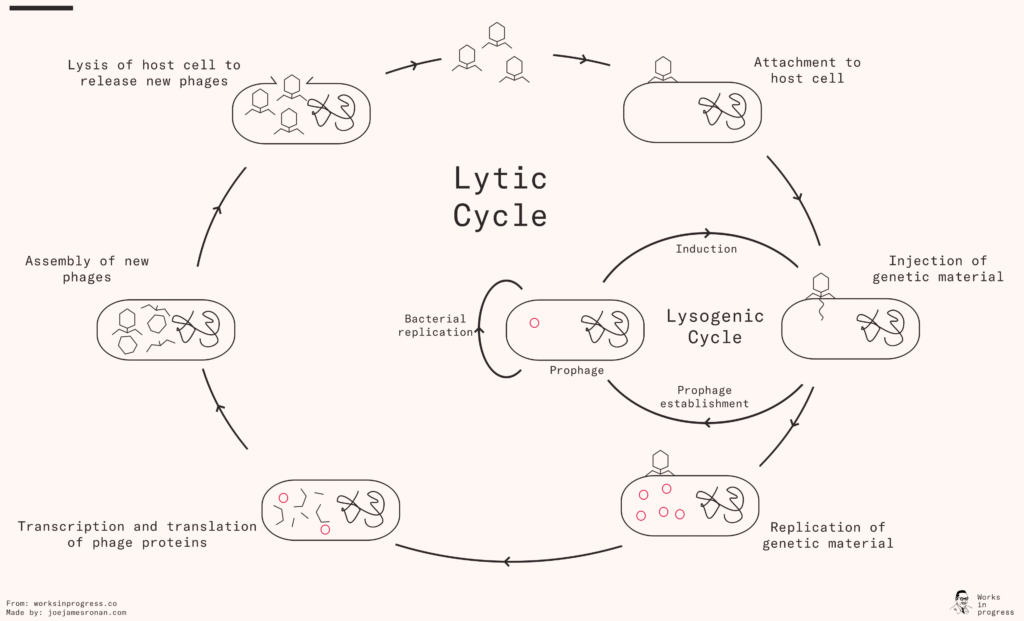
During the lytic cycle, the phage hijacks the bacterial host to replicate its genetic material and produce the proteins it needs for its capsule. When they are assembled, the phage causes the bacterial cell to disintegrate – a process called lysis, hence also the name of the two cycles (lytic and lysogenic) – which releases the new phages into the environment, and the cycle can begin anew.
Phages are the most abundant biological entity on the planet.
It’s estimated that one milliliter of seawater contains around ten million phages. In recent years, we have learned a lot about the human microbiome, which encompasses all the trillions of invisible microorganisms that live in and on our body, many with symbiotic relationships with humans. One thing we have learnt is that the human body is also colonized by a huge number of phages – there are ten times more of them than all other microorganisms put together.
This is not all good news for humans. As phages kill bacteria, bacteria evolve in response. In some cases, the prophage lying dormant inside a bacterium can make that bacterium more dangerous. For example, Vibrio cholerae, the bacterium that causes cholera, is only pathogenic when infected with a phage called CTXφ, which encodes the cholera toxin in its genome.
Using phages as therapeutic agents is not a new idea. Félix d’Hérelle recognized the therapeutic potential of phages, describing the correlation between the presence of phages and the clearing of diseases in his first paper in 1917. He isolated the ‘anti microbe’ from stool samples of patients who had recovered from dysentery. He added 4-5 drops of stool to a nutritious broth and incubated it for 18 hours at 37°C. To ensure that there were no bacteria present in the sample, he filtered it using a ceramic filter called Chamberland filter. A few drops of the bacterium-free filtrates were sufficient to kill a culture of Shigella. To further test the potential of this isolated phage to work as a therapy, d’Hérelle performed an experiment in rabbits, where he was able to show that phages could protect from a Shigella infection, although details about this study are difficult to find today.
The first time phage therapy was applied in humans was in Paris in 1919, after d’Hérelle and several hospital workers swallowed the phage preparation to confirm its safety. They then administered the therapy to a 12-year-old boy who suffered from severe dysentery. A single administration was sufficient to get rid of his symptoms and he recovered fully within a few days. D’Hérelle even started a commercial laboratory in Paris, which produced therapeutic phages against various bacterial infections, and which later became l’Oréal.
When antibiotics were discovered, they led to a decline in interest in phage therapy, since antibiotics were far easier to deploy against a wide variety of infections, whereas phages had to be much more targeted towards particular strains of bacteria. Only the Soviet Union and some Eastern European countries continued to use phages, due to their limited access to antibiotics. The Georgi Eliava Institute in Tbilisi, Georgia, has been doing phage research and phage therapy since 1923 and has the most extensive collection of bacterial strains and phages in the world.
Phage therapy has several advantages compared to antibiotics. Firstly and most importantly, there are phages for multi-resistant bacterial strains. They are also unable to harm not just the human host of the bacteria, but even the ‘good bacteria’ of the broader microbiome, as they can only infect very specific bacteria with a matching keyhole. Thirdly, they are able to multiply as long as their bacterial hosts are present, which leads to a self-dosing therapy.
Unfortunately, their specificity can also be a disadvantage. To prescribe a phage for a patient, you would need to precisely identify the strain that causes their disease, or else the bacteria will not be infected and killed. In many cases, we have only a vague idea of what bacteria is infecting someone – perhaps just whether it is Gram-positive or -negative, a simple classification of the type of cell wall that bacteria have. Ideally, we would be able to culture a sample in a dish and then sequence its genome, to confirm which one was inside our patient, and use phages that could specifically target that bacteria. But without it, our phages would be unlikely to do anything.
But there is another solution: phage cocktails, which are therapies that include multiple phages. One approach is to design a cocktail for a specific patient. This customized approach may take weeks. It means isolating the infectious agent then testing it against a phage library to narrow down the selection of phages for the cocktail. But you could also develop a phage cocktail with less information. By knowing about where in the body the infection is, or the symptoms, you could rule out some species and make a better guess about which species might be responsible for the infection. The cocktail would then include phages specific for the species commonly causing this disease.
And there’s the other disadvantage we talked about before. Bacteria can evolve resistance to phages – even during the course of a single treatment. Bacteria and phages are in an evolutionary arms race, adapting to survive when the other gains an upper hand. And since phages are taken from the environment, usually from human sewage, some bacteria may already have some resistance before treatment has begun.
This means some phage mixtures may need to be updated: the ineffective ones can either be replaced by new phages or ‘trained’. This involves putting pressure onto phages, which speeds up their rate of evolution – by culturing them with phage-resistant bacteria, only those that have mutations that let them remain active can survive.
Like most other viruses, phages that kill every single one of their hosts before they can spread to others tend to die out rapidly. Phages in nature are more successful if they control their bacterial host population without wiping it out completely. For humans, therefore, phages may work best as a biocontrol agent, reducing the sheer numbers of pathogens, and giving the human immune system (or antibiotics) a chance to eliminate the remaining bacteria. In this case, they may be used as a complement to antibiotics, to treat milder infections without risking further antibiotic resistance.
With a more vibrant phage research ecosystem, these and other questions can be answered. The most important concern is how they evolve and how resistances against them develop. Antibiotics have been misused and overused. The same thing must be avoided with phages, because applying them too broadly could cause rapid resistance and put us back to square one.
But there are other options too. We don’t necessarily need to use entire phages to fight against bacteria. Phages could also be the source for individual proteins used in simpler chemical drugs.
For example, we know that while phages reproduce in the lytic cycle, they produce endolysins, proteins that cause cell lysis: where bacterial cells burst and release newly produced phages. In recent years, scientists have discovered that endolysins are effective antimicrobials against resistant strains of Staphylococcus aureus and Listeria monocytogenes. So far, we have not seen any resistance forming to endolysins, even in lab experiments deliberately designed to generate it. As well as this, endolysins can kill dormant bacteria that are hiding inside human cells or protected by biofilms, which antibiotics are unable to do.
Endolysins have a further advantage: their structure is quite simple. This means that we can produce them in a lab, and can even edit them or design new ones. But they currently have some limitations as well. At the moment they are more suitable for targeting Gram-positive bacteria – Gram-negative bacteria have an additional outer membrane protecting the cell wall from endolysin attacks.
But the downsides seem solvable. For example, it can be difficult to deliver endolysins into specific tissues, like the heart tissue in the case of endocarditis, or into specific cells, where intracellular bacteria reside. But we can fuse them with peptides that penetrate cells or home into specific tissues to increase the drug concentration at the target site and help cells to take up the antimicrobial. This approach has been successful in cell lines and a mouse model for abscesses. Other challenges arise from their composition: because they are foreign proteins, they can trigger an immune response against them.
We can overcome some of these challenges too. In order to increase their lifespans, scientists have fused endolysins to a specific domain leading to reduced renal clearance and degradation. In a systemic mouse model, this change led to better therapeutic outcomes compared to the original endolysin.
N-Rephasin SAL200 and CF-301 are two endolysins in clinical trials that might help patients suffering bacteremia from Staphylococcus aureus. Both are administered intravenously and in combination with standard antibiotic treatment, and the results of CF-301 show that patients responded better to the combined treatment than if they were treated with antibiotics alone.
The state of play
Phages are already saving lives.
A 30-year-old woman needed extensive surgery after being a victim of the suicide bombing at Brussels Airport in 2016. Four days after the surgery, she had septic shock due to an infection of the surgical wound. After 700 days of antibiotic treatments and various surgeries, she was treated with a cocktail of bacteriophages as well as more antibiotics, which was injected via a catheter directly into the infected wound. After this, her condition finally improved, and her infection was eliminated.
But phage therapy is limited by our knowledge of different phages and current drug approval regimes, which aren’t designed smoothly for the huge number of possible phages we might want to use.
There are at least as many potential types of phages as there are pathogenic bacterial strains – and then as many cocktails as there are potential combinations. This adds up to trillions and trillions of possibilities, many quite specific to narrow situations, and many requiring updating over time. But the Federal Drug Administration, which approves drugs in the USA, by far the world’s biggest drug market, treats each phage cocktail as a new entity.
This means that each cocktail would need to go through perhaps a decade of Phase I, II, and III trials, including randomized controlled trials on tens of thousands of people. If each one of these uncountable cocktails has only a relatively small market, then they are unlikely to be worth the millions of dollars these trials cost.
By contrast, agents used for food safety and agriculture do not require such extensive pre-approval, and phages are already used in these fields. Listex P100 (Micreos; the Netherlands) and ListShield (Intralytix; Baltimore, Maryland, USA) are two examples of approved and commercially available phage products to protect processed food from Listeria monocytogenes.
There is great potential in bacteriophages and phage-derived products to tackle the antibiotic crisis. Given the technology as it exists today, they could be used in combination with standard antibiotics to treat bacterial infections and help us reduce our dependence on existing antibiotics, reducing the growth rate of bacterial resistance. But given further research and development, they have the potential to go well beyond that in treating disease.
Being aware of how phages work and their potential might help to address the regulatory obstacles currently in the way. Phages and phage products need to be treated in a more dynamic way, similar to the yearly adaptation of the flu shot. There is no fundamental reason we could not do that, as long as we get the chance to try.